Human exonic regions account for only 1-2% of the whole genome, yet they contain about 85% of disease-causing mutations. Whole exome sequencing (WES) can significantly improve the detection rate of low-frequency mutations and rare variants, especially in the fields of prenatal screening and diagnosis, genetic diseases, and tumor minimal residual disease (MRD) research and application.
GeneMind has collaborated with Agilent to provide a WES total solution through Agilent’s SureSelect V8 capture and library preparation kits, based on GeneMind’s GenoLab M, FASTASeq 300, and SURFSeq 5000 sequencing platforms. Test results demonstrate high compatibility between GeneMind’s sequencing platforms and Agilent’s WES library preparation kits, with high specificity, a high mapping rate, uniform genome coverage, a high on-target ratio, and consistent variant detection.
GeneMind & Agilent WES Solution Workflow

High Data Quality
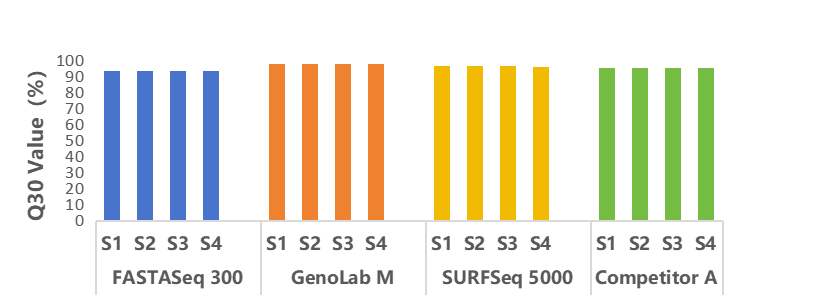
Four libraries were constructed using Agilent’s SureSelect V8. Each library was then sequenced and analyzed on GeneMind’s FASTASeq 300, GenoLab M, and SURFSeq 5000 platforms, as well as on a competitor’s platform (Competitor A). The sequencing results showed that the Q30 values of the data from GeneMind’s platforms and Competitor A’s platform were both over 93%, with no significant difference. This ensures the accuracy and reliability of the sequencing results.
Validation Results
(1)High mapping rate and on-target ratio
The genome mapping rate of all samples was close to 100%, the on-target ratio was more than 86%, and the 30X target region coverage rate was more than 97.7%.
| Samples-S1 | FASTASeq 300 | GenoLab M | SURFSeq 5000 | Competitor A |
| Total reads | 85187632 | 79835224 | 79556138 | 79160548 |
| Mapped reads | 85171990 | 79824734 | 79540935 | 79135311 |
| Map ratio | 99.98% | 99.99% | 99.98% | 99.97% |
| Dup ratio | 9.63% | 8.46% | 14.59% | 30.22% |
| Percent of paired reads on target region | 88.11% | 87.98% | 86.78% | 86.59% |
| Mean depth | 171.75 | 162.94 | 149.7 | 121.7 |
| Percent of target base covered at least 30X | 97.75% | 97.79% | 97.79% | 97.62% |
| Samples-S2 | FASTASeq 300 | GenoLab M | SURFSeq 5000 | Competitor A |
| Total reads | 84000130 | 79834398 | 79595356 | 90612170 |
| Mapped reads | 83981840 | 79823942 | 79579812 | 90583279 |
| Map ratio | 99.98% | 99.99% | 99.98% | 99.97% |
| Dup ratio | 9.68% | 8.63% | 16.96% | 33.05% |
| Percent of paired reads on target region: | 88.02% | 87.89% | 86.69% | 86.57% |
| Mean depth | 169.56 | 162.89 | 145.77 | 133.86 |
| Percent of target base covered at least 30X | 97.73% | 97.82% | 97.77% | 97.89% |
| Samples-S3 | FASTASeq 300 | GenoLab M | SURFSeq 5000 | Competitor A |
| Total reads | 87507016 | 79844860 | 79578274 | 88721930 |
| Mapped reads | 87493322 | 79835097 | 79563465 | 88693939 |
| Map ratio | 99.98% | 99.99% | 99.98% | 99.97% |
| Dup ratio | 9.48% | 8.18% | 15.11% | 31.14% |
| Percent of paired reads on target region: | 88.30% | 88.16% | 87.02% | 86.93% |
| Mean depth | 177.6 | 164.22 | 149.59 | 135.34 |
| Percent of target base covered at least 30X | 97.77% | 97.76% | 97.74% | 97.84% |
| Samples-S4 | FASTASeq 300 | GenoLab M | SURFSeq 5000 | Competitor A |
| Total reads | 89651756 | 79839616 | 79536922 | 90201704 |
| Mapped reads | 89634832 | 79828842 | 79519977 | 90168637 |
| Map ratio | 99.98% | 99.99% | 99.98% | 99.96% |
| Dup ratio | 9.55% | 7.99% | 14.09% | 31.43% |
| Percent of paired reads on target region: | 87.93% | 87.80% | 86.49% | 86.22% |
| Mean depth | 180.99 | 163.84 | 150.17 | 135.82 |
| Percent of target base covered at least 1X | 99.64% | 99.63% | 99.63% | 99.63% |
| Percent of target base covered at least 10X | 99.19% | 99.18% | 99.17% | 99.21% |
| Percent of target base covered at least 30X | 97.90% | 97.84% | 97.82% | 97.95% |
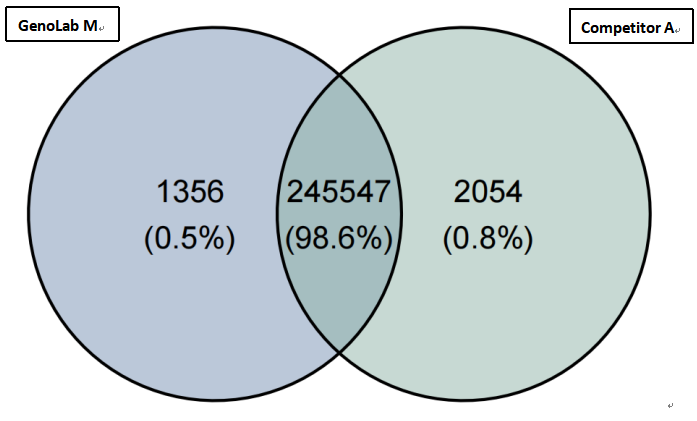
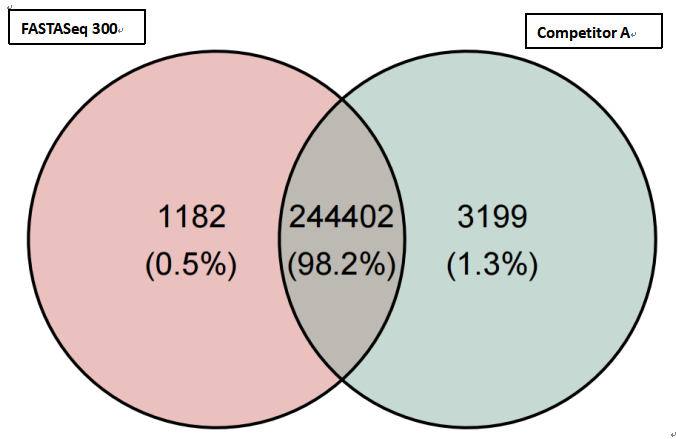
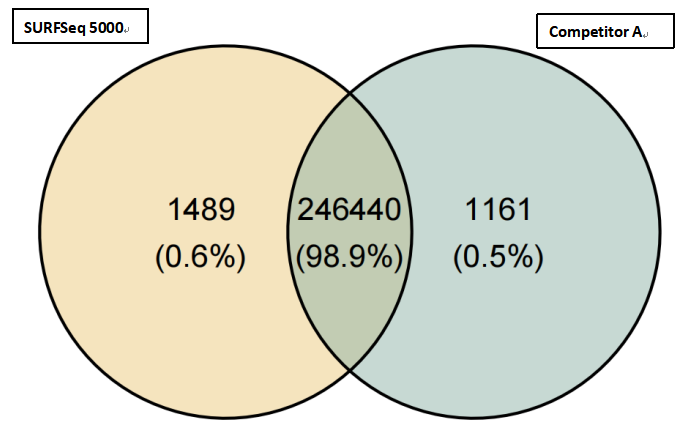
(2)High consistency of the variant site detection
The consistency of SNV/InDel detection between GeneMind’s FASTASeq 300 (FA), GenoLab M (GL), and SURFSeq 5000 (SF) platforms showed a highly comparable result with Competitor A. In all samples, SNV/InDel detection rates exceeded 98.2%, demonstrating a very high level of consistency and ensuring the accuracy and reliability of variant site detection.
*Unless otherwise informed, GeneMind sequencing platform and related sequencing reagents are not available in the USA, Canada, Australia, Japan, Singapore, Western Europe, and Nordic countries yet.




